investigating unstudied metalloenzyme families
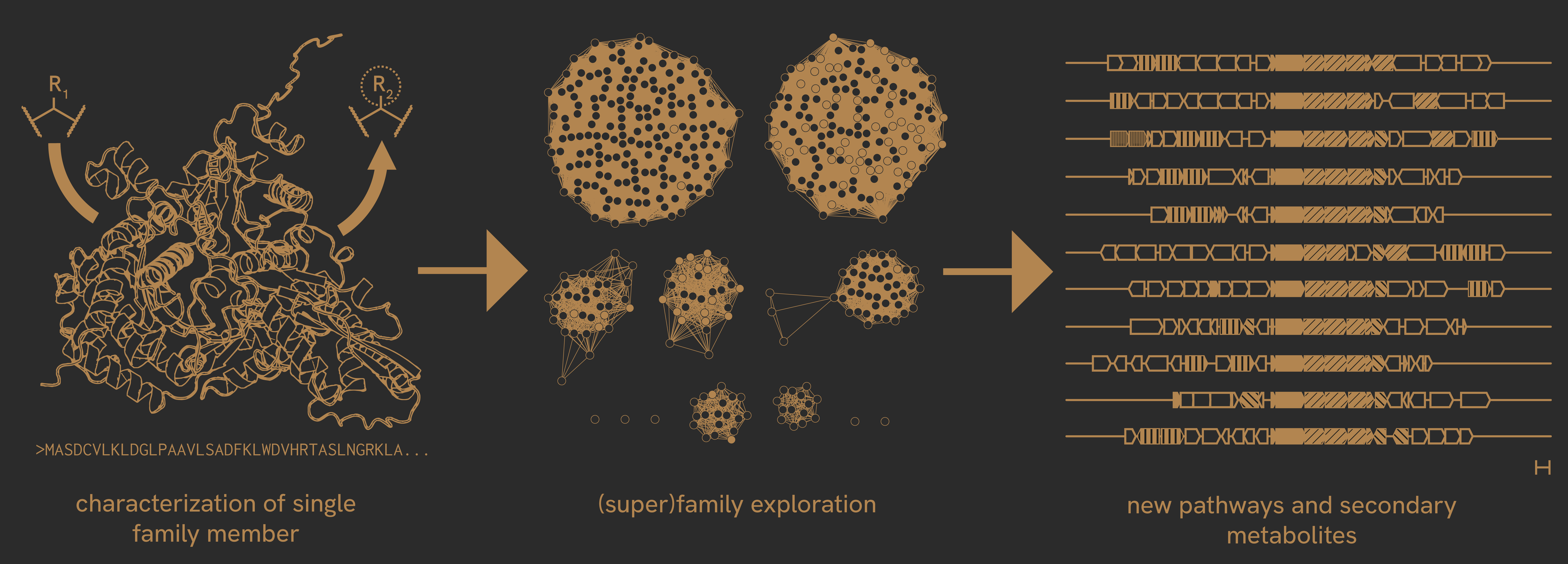
How we can identify and explore the new protein families that are being identified in microbial genomic dark matter? Many un- or understudied metalloenzyme superfamilies can carry out diverse complex biochemical reactions. Dr. Kenney's past in silico, in vitro and in vivo work has identified several new enzyme (sub)families which have been prioritized for further investigation - in particular, enzyme classes involved in new heteroatomic post-translational modifications of amino acids. In this project, we will explore the biochemistry of these enzymes as well as the roles of their natural products in microbial cells. More broadly, we will use this project as a testing ground for developing computational tools for hypothesis generation and prioritization of novel microbial enzymes.
exploring poorly understood enzyme superfamilies
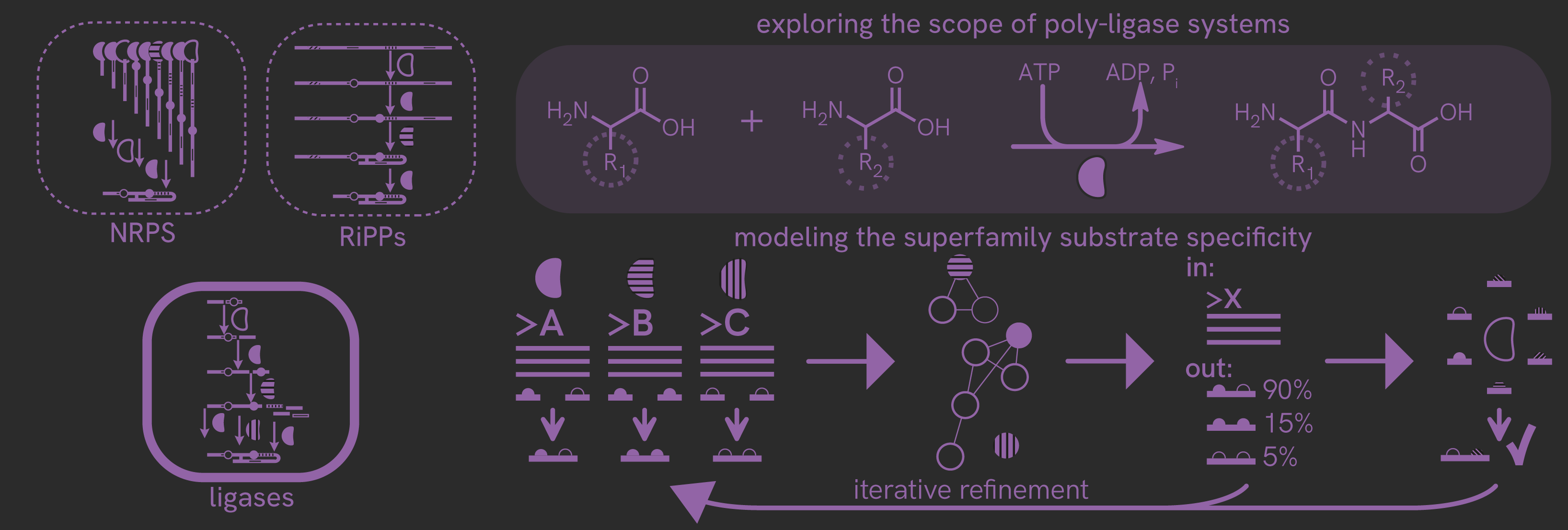
How can we put assemble reactivity rules for enzyme superfamilies using information about individual enzymes? This project focuses on an underappreciated enzyme class that provides a route towards the production of oligopeptidic natural products distinct from the well-established NRPS and RiPP pathways. A computational model tying sequence and structure to substrate reactivity will be established, and will be experimentally validated and expanded as we investigate new family members and the natural products they produce. As a long term goal, we hope to use members of this enzyme family for biocatalysis and for the assembly of "non-natural" products.
discovering metallophores in complex microbial communities
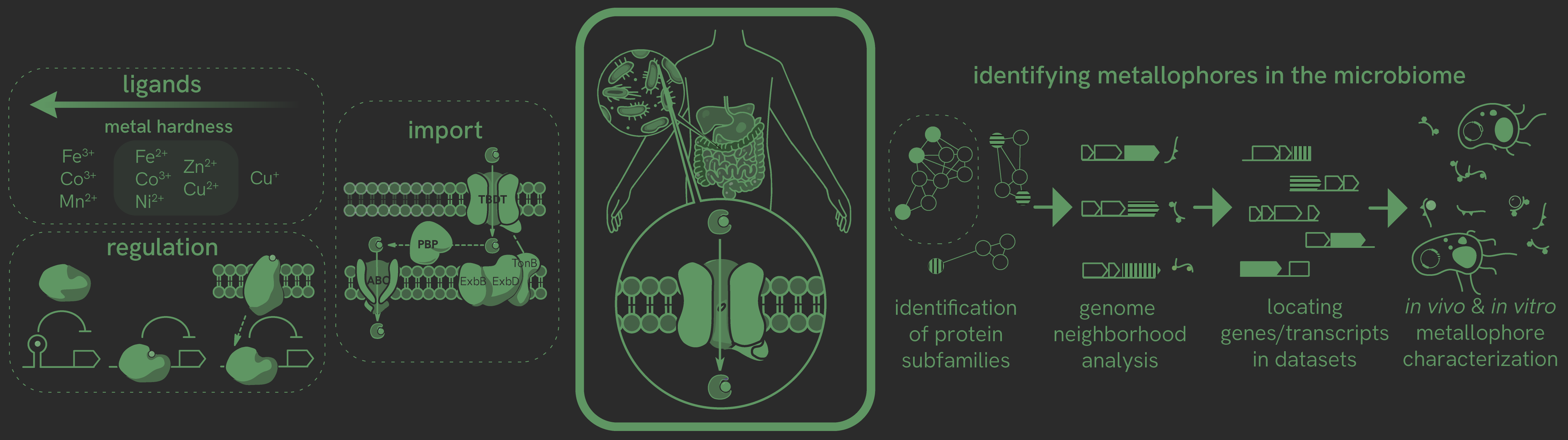
How can we explore chemistry that we know microbes are probably carrying out, but that hasn't been tied to any known pathway? Research into microbial metal acquisition in human hosts has focused on host-pathogen competition for ferric iron. However, it is now clear that metal-binding natural products involved in metal uptake exist for many metals. We hypothesize that unique metallophores exist in the human gut microbiome, which is an anaerobic and densely populated environment distinct from those in which metallophores have been best studied. Bioinformatics, metagenomics, and metatranscriptomics will guide our identification of these metallophores, and new compounds will be structurally and (bio)chemically characterized. This will answer fundamental questions about microbial physiology and have important implications for human health.